Search
- Page Path
- HOME > Search
-
Evaluation of mineral induction ability and cytotoxicity of carbonated hydroxyapatite for pulp tissue regeneration: an
in vitro study - S. Swathi Priyadharshini, Chinnasamy Ragavendran, Anand Sherwood, J. Ramana Ramya, Jogikalmat Krithikadatta
- Restor Dent Endod 2024;49(4):e40. Published online October 29, 2024
- DOI: https://doi.org/10.5395/rde.2024.49.e40
-
 Abstract
Abstract
 PDF
PDF PubReader
PubReader ePub
ePub Objectives This study aimed to evaluate carbonated hydroxyapatite (CHA)’s ability for mineral induction and its
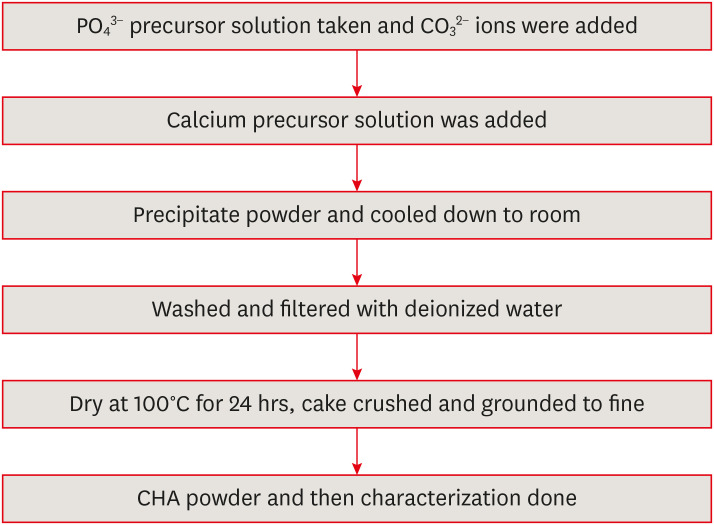
in vitro cytotoxicity with human dental pulp cells.Materials and Methods Precursors for the study include di-ammonium hydrogen phosphate and calcium nitrate tetrahydrate, with sodium hydrogen carbonate added to achieve different levels of carbonate substitution. The synthesized CHA samples are characterized using X-ray diffraction, Fourier transform infrared spectroscopy, and Raman spectroscopy. Scanning electron microscopy (SEM) was used to observe morphology. For 14 days at 37°C, samples were submerged in simulated body fluid to assess their mineral induction capabilities. SEM was used to confirm apatite formation on sample surfaces. The cytotoxicity assay was used to assess the vitality of the cells following their exposure to various concentrations of CHA.
Results The Joint Committee on Powder Diffraction Standards data for HA aligned well with the results from X-ray diffraction analysis of CHA across 3 different concentrations, indicating strong agreement. Fourier transform infrared spectra indicated the presence of phosphate, hydroxyl, and carbonate groups within the samples. SEM and Energy-dispersive X-ray analysis show agglomerated and flaky nanoparticles. All the samples are bioactive, but the formation of apatite differs from one another.
In vitro cytotoxicity assay showed that over 70% of cells maintain viability.Conclusions The results of this study may provide insight into the potential use of carbonated HA as a dental pulp-capping material for vital pulp therapy.
-
Citations
Citations to this article as recorded by- Smart Nanomaterials: Current State and Future Prospects in Drug Delivery and Tissue Engineering
E. Elizabeth Rani, D. Sakthi Sanjana, E. Karthikeyan, J. Nandhini
Biomedical Materials & Devices.2026; 4(2): 1455. CrossRef - Physicochemical and antibacterial evaluation of novel nano α-TCP–AgNPs biocomposites for direct pulp-capping applications
Selviana Wulansari, Hendra Dian Adhita Dharsono, Nasrul Wathoni, Rosalina Tjandrawinata, Arief Cahyanto, Moehamad Orliando Roeslan
Frontiers in Oral Health.2026;[Epub] CrossRef - Physicochemical effects of nano type-B bone substitute on pulp protective cement formulations
Njwan Fadhel SHEHAB
Dental Materials Journal.2026; 45(1): 92. CrossRef - Comparative evaluation of compressive strength and morphological interface of carbonated hydroxyapatite with other pulp capping materials: An in vitro analysis
S. Swathi Priyadharshini, Chinnasamy Ragavendran, I. Anand Sherwood, Ramanaramya Jeyapalan
Endodontology.2025; 37(1): 90. CrossRef - Thermoresponsive Nanomaterials: Revolutionizing Cancer Theranostics
Bellarmin Michael, Mohanakrishnan Srinivasan, Karthikeyan Elumalai, Lokeshwar Ravikumar, Sivaprakash Kathiresan, Nandhini Jayaprakash
Biomedical Materials & Devices.2025;[Epub] CrossRef - Bioactive Dioxo-Phosphobetaines derived from the reaction of Dichlorodinitrobenzofuroxane with various phosphines
Irina V. Galkina, Haiyan Fan, Semen R. Romanov, Dmitriy I. Bakhtiyarov, Luisa M. Usupova, Svetlana N. Egorova, Yulia V. Bakhtiyarova, Enrico Benassi
Bioorganic Chemistry.2025; 163: 108695. CrossRef - Near-infrared laser-activated PLGA-PDA core-shell nanohybrids for synergistic photothermal antibacterial therapy and sustained ion release in orthodontic white spot lesions prevention
Zezhou Feng, Yujiang Liu, Silu Sun, Minmin Si, Di Huang, Zhiyuan Feng
Journal of Dentistry.2025; 162: 106078. CrossRef - Formation and utilization of soluble microbial products in denitrifying biofilters at different carbon-to-nitrogen ratios: Microbial community characteristics
Fangyuan Jiang, Xianyang Shi
Journal of Environmental Chemical Engineering.2025; 13(6): 119554. CrossRef - Bioactivity and biocompatibility of bioceramic-based pulp capping materials in laboratory and animal models
Rafiqul Islam, Md. Refat Readul Islam, Kenta Tsuchiya, Yu Toida, Hidehiko Sano, Monica Yamauti, Hany Mohamed Aly Ahmed, Atsushi Tomokiyo
Journal of Materials Science: Materials in Medicine.2025;[Epub] CrossRef - Physical, Chemical, and Biological Properties of Graphene Nanoparticle-added Tricalcium Silicate Formulations: A Systematic Review
Soundaria Srinivasan, Deepa Gurunathan, Lakshmi Thangavelu
Journal of International Oral Health.2025; 17(6): 453. CrossRef - Advanced structural and compositional profiling of mineral trioxide aggregate incorporated with nano-carbonated hydroxyapatite: a comprehensive X-ray diffraction and energy dispersive X-ray investigation
Njwan Fadhel Shehab, Nadia Hameed Hasan, Alaa Edrees Dawood, Nawal Atiya Khalaf
Biomaterial Investigations in Dentistry.2025; 12: 216. CrossRef
- Smart Nanomaterials: Current State and Future Prospects in Drug Delivery and Tissue Engineering
- 2,918 View
- 126 Download
- 9 Web of Science
- 11 Crossref

- The comparison of gene expression from human dental pulp cells and periodontal ligament cells
- Hyoun So, Sang-Hyuk Park, Gi-Woon Choi
- J Korean Acad Conserv Dent 2009;34(5):430-441. Published online September 30, 2009
- DOI: https://doi.org/10.5395/JKACD.2009.34.5.430
-
 Abstract
Abstract
 PDF
PDF PubReader
PubReader ePub
ePub The purpose of this study was to characterize functional distinction between human dental pulp cells(PC) and periodontal ligament cells(PDLC) using cDNA microarray assay and to confirm the results of the microarray assay using RT-PCR. 3 genes out of 51 genes which were found to be more expressed(>2 fold) in PC were selected, and 3 genes out of 19 genes which were found to be more expressed(>2 fold) in PDLC were selected for RT-PCR as well.
According to this study, the results were as follows:
1. From the microarray assay, 51 genes were more expressed (2 fold) from PC than PDLC.
2. RT-PCR confirmed that ITGA4 and TGF β2 were more expressed in PC than in PDLC.
3. From the microarray assay, 19 genes were more expressed (2 fold) from PDLC than PC.
4. RT-PCR confirmed that LUM, WISP1, and MMP1 were more expressed in PDLC than in PC.
From the present study, different expression of the genes between the PC and PDLC were characterized to show the genes which play an important role in dentinogenesis were more expressed from PC than PDLC, while the genes which were related with collagen synthesis were more expressed from PDLC than PC.
-
Citations
Citations to this article as recorded by- Gene expression profiling in human dental pulp cells treated with mineral trioxide aggregate
Yong-Beom Kim, Won-Jun Shon, WooCheol Lee, Kee-Yeon Kum, Seung-Ho Baek, Kwang-Shik Bae
Journal of Korean Academy of Conservative Dentistry.2010; 35(3): 152. CrossRef
- Gene expression profiling in human dental pulp cells treated with mineral trioxide aggregate
- 1,110 View
- 1 Download
- 1 Crossref

- Tissue engineering of dental pulp on type I collagen
- Gwang-Hee Lee, Sung-Yoon Huh, Sang-Hyuk Park
- J Korean Acad Conserv Dent 2004;29(4):370-377. Published online July 31, 2004
- DOI: https://doi.org/10.5395/JKACD.2004.29.4.370
-
 Abstract
Abstract
 PDF
PDF PubReader
PubReader ePub
ePub The purpose of this study was to regenerate human dental pulp tissues similar to native pulp tissues. Using the mixture of type I collagen solution, primary cells collected from the different tissues (pulp, gingiva, and skin) and NIH 3T3 (1 × 105 cells/ml/well) were cultured at 12-well plate at 37℃ for 14 days. Standardized photographs were taken with digital camera during 14 days and the diameter of the contracted collagen gel matrix was measured and statistically analyzed with student t-test. As one of the pulp tissue engineering, normal human dental pulp tissue and collagen gel matrix cultured with dental pulp cells for 14 days were fixed and stained with Hematoxyline & Eosin.
According to this study, the results were as follows:
1. The contraction of collagen gel matrix cultured with pulp cells for 14 days was significantly higher than other fibroblasts (gingiva, skin) (p < 0.05).
2. The diameter of collagen gel matrix cultured with pulp cells was reduced to 70.4% after 7 days, and 57.1% after 14 days.
3. The collagen gel without any cells did not contract, whereas the collagen gel cultured with gingiva and skin showed mild contraction after 14 days (88.1% and 87.6% respectively).
4. The contraction of the collagen gel cultured with NIH 3T3 cells after 14 days was higher than those cultured with gingival and skin fibroblasts, but it was not statistically significant (72.1%, p > 0.05).
5. The collagen gel matrix cultured with pulp cells for 14 days showed similar shape with native pulp tissue without blood vessels.
This approach may provide a means of engineering a variety of other oral tissue as well and these cell behaviors may provide information needed to establish pulp tissue engineering protocols.
-
Citations
Citations to this article as recorded by- Human amniotic membrane extracellular matrix scaffold for dental pulp regeneration in vitro and in vivo
Hengameh Bakhtiar, Azin Ashoori, Sarah Rajabi, Mohamad Pezeshki‐Modaress, Alireza Ayati, Mohammad Reza Mousavi, Mohammad Reza Ellini, Amir Kamali, Amir Azarpazhooh, Anil Kishen
International Endodontic Journal.2022; 55(4): 374. CrossRef
- Human amniotic membrane extracellular matrix scaffold for dental pulp regeneration in vitro and in vivo
- 1,582 View
- 2 Download
- 1 Crossref


 KACD
KACD

 First
First Prev
Prev