Search
- Page Path
- HOME > Search
-
Evaluation of mineral induction ability and cytotoxicity of carbonated hydroxyapatite for pulp tissue regeneration: an
in vitro study - S. Swathi Priyadharshini, Chinnasamy Ragavendran, Anand Sherwood, J. Ramana Ramya, Jogikalmat Krithikadatta
- Restor Dent Endod 2024;49(4):e40. Published online October 29, 2024
- DOI: https://doi.org/10.5395/rde.2024.49.e40
-
 Abstract
Abstract
 PDF
PDF PubReader
PubReader ePub
ePub Objectives This study aimed to evaluate carbonated hydroxyapatite (CHA)’s ability for mineral induction and its
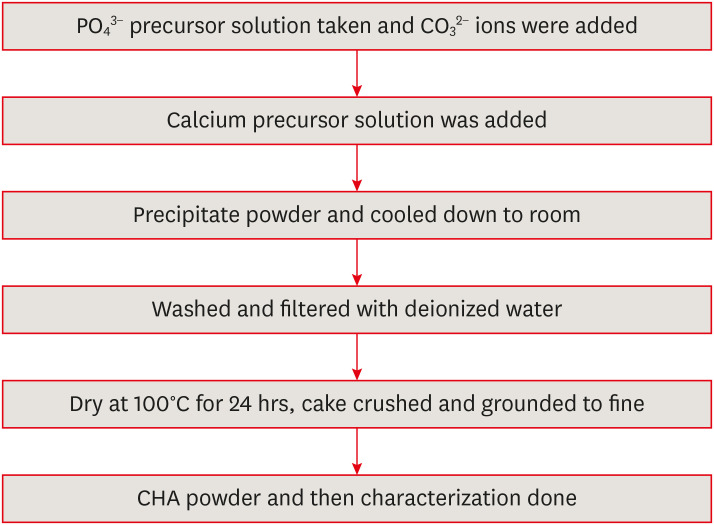
in vitro cytotoxicity with human dental pulp cells.Materials and Methods Precursors for the study include di-ammonium hydrogen phosphate and calcium nitrate tetrahydrate, with sodium hydrogen carbonate added to achieve different levels of carbonate substitution. The synthesized CHA samples are characterized using X-ray diffraction, Fourier transform infrared spectroscopy, and Raman spectroscopy. Scanning electron microscopy (SEM) was used to observe morphology. For 14 days at 37°C, samples were submerged in simulated body fluid to assess their mineral induction capabilities. SEM was used to confirm apatite formation on sample surfaces. The cytotoxicity assay was used to assess the vitality of the cells following their exposure to various concentrations of CHA.
Results The Joint Committee on Powder Diffraction Standards data for HA aligned well with the results from X-ray diffraction analysis of CHA across 3 different concentrations, indicating strong agreement. Fourier transform infrared spectra indicated the presence of phosphate, hydroxyl, and carbonate groups within the samples. SEM and Energy-dispersive X-ray analysis show agglomerated and flaky nanoparticles. All the samples are bioactive, but the formation of apatite differs from one another.
In vitro cytotoxicity assay showed that over 70% of cells maintain viability.Conclusions The results of this study may provide insight into the potential use of carbonated HA as a dental pulp-capping material for vital pulp therapy.
-
Citations
Citations to this article as recorded by- Comparative evaluation of compressive strength and morphological interface of carbonated hydroxyapatite with other pulp capping materials: An in vitro analysis
S. Swathi Priyadharshini, Chinnasamy Ragavendran, I. Anand Sherwood, Ramanaramya Jeyapalan
Endodontology.2025; 37(1): 90. CrossRef - Smart Nanomaterials: Current State and Future Prospects in Drug Delivery and Tissue Engineering
E. Elizabeth Rani, D. Sakthi Sanjana, E. Karthikeyan, J. Nandhini
Biomedical Materials & Devices.2025;[Epub] CrossRef - Thermoresponsive Nanomaterials: Revolutionizing Cancer Theranostics
Bellarmin Michael, Mohanakrishnan Srinivasan, Karthikeyan Elumalai, Lokeshwar Ravikumar, Sivaprakash Kathiresan, Nandhini Jayaprakash
Biomedical Materials & Devices.2025;[Epub] CrossRef - Bioactive Dioxo-Phosphobetaines derived from the reaction of Dichlorodinitrobenzofuroxane with various phosphines
Irina V. Galkina, Haiyan Fan, Semen R. Romanov, Dmitriy I. Bakhtiyarov, Luisa M. Usupova, Svetlana N. Egorova, Yulia V. Bakhtiyarova, Enrico Benassi
Bioorganic Chemistry.2025; 163: 108695. CrossRef - Near-infrared laser-activated PLGA-PDA core-shell nanohybrids for synergistic photothermal antibacterial therapy and sustained ion release in orthodontic white spot lesions prevention
Zezhou Feng, Yujiang Liu, Silu Sun, Minmin Si, Di Huang, Zhiyuan Feng
Journal of Dentistry.2025; 162: 106078. CrossRef - Formation and utilization of soluble microbial products in denitrifying biofilters at different carbon-to-nitrogen ratios: Microbial community characteristics
Fangyuan Jiang, Xianyang Shi
Journal of Environmental Chemical Engineering.2025; 13(6): 119554. CrossRef - Bioactivity and biocompatibility of bioceramic-based pulp capping materials in laboratory and animal models
Rafiqul Islam, Md. Refat Readul Islam, Kenta Tsuchiya, Yu Toida, Hidehiko Sano, Monica Yamauti, Hany Mohamed Aly Ahmed, Atsushi Tomokiyo
Journal of Materials Science: Materials in Medicine.2025;[Epub] CrossRef - Physical, Chemical, and Biological Properties of Graphene Nanoparticle-added Tricalcium Silicate Formulations: A Systematic Review
Soundaria Srinivasan, Deepa Gurunathan, Lakshmi Thangavelu
Journal of International Oral Health.2025;[Epub] CrossRef - Advanced structural and compositional profiling of mineral trioxide aggregate incorporated with nano-carbonated hydroxyapatite: a comprehensive X-ray diffraction and energy dispersive X-ray investigation
Njwan Fadhel Shehab, Nadia Hameed Hasan, Alaa Edrees Dawood, Nawal Atiya Khalaf
Biomaterial Investigations in Dentistry.2025; 12: 216. CrossRef
- Comparative evaluation of compressive strength and morphological interface of carbonated hydroxyapatite with other pulp capping materials: An in vitro analysis
- 2,342 View
- 117 Download
- 7 Web of Science
- 9 Crossref

- Changes in SIRT gene expression during odontoblastic differentiation of human dental pulp cells
- Young-Eun Jang, Su-Hee Go, Bin-Na Lee, Hoon-Sang Chang, In-Nam Hwang, Won-Mann Oh, Yun-Chan Hwang
- Restor Dent Endod 2015;40(3):223-228. Published online July 15, 2015
- DOI: https://doi.org/10.5395/rde.2015.40.3.223
-
 Abstract
Abstract
 PDF
PDF PubReader
PubReader ePub
ePub Objectives The aim of this study was to investigate the expression of 7 different sirtuin genes (SIRT1-SIRT7) in human dental pulp cells (HDPCs), and to determine the role of SIRTs in the odontoblastic differentiation potential of HDPCs.
Materials and Methods HDPCs were isolated from freshly extracted third molar teeth of healthy patients and cultulred in odontoblastic differentiation inducing media. Osteocalcin (OCN) and dentin sialophosphoprotein (DSPP) expression was analyzed to evaluate the odontoblastic differentiation of HDPCs by reverse transcription-polymerase chain reaction (RT-PCR), while alizarin red staining was used for the mineralization assay. To investigate the expression of SIRTs during odontoblastic differentiation of HDPCs, real time PCR was also performed with RT-PCR.
Results During the culture of HDPCs in the differentiation inducing media, OCN, and DSPP mRNA expressions were increased. Mineralized nodule formation was also increased in the 14 days culture. All seven SIRT genes were expressed during the odontogenic induction period. SIRT4 expression was increased in a time-dependent manner.
Conclusions Our study identified the expression of seven different SIRT genes in HDPCs, and revealed that SIRT4 could exert an influence on the odontoblast differentiation process. Further studies are needed to determine the effects of other SIRTs on the odontogenic potential of HDPCs.
-
Citations
Citations to this article as recorded by- Biodegradable Zn‐5Dy Alloy with Enhanced Osteo/Angio‐Genic Activity and Osteointegration Effect via Regulation of SIRT4‐Dependent Mitochondrial Function
Yue Han, Xian Tong, Runqi Zhou, Yilin Wang, Yuge Chen, Liang Chen, Xinhua Hong, Linmei Wu, Zhiqiang Lin, Yichi Zhang, Xuejia Zhang, Chaoming Hu, Bin Li, Yifan Ping, Zelin Cao, Zhou Ye, Zhongchen Song, Yuncang Li, Cuie Wen, Yongsheng Zhou, Jixing Lin, Shen
Advanced Science.2024;[Epub] CrossRef - The Role of Histone Acetylation Modification in Dental Tissue-Derived Mesenchymal Stem Cells and Odontogenesis
Haoling Chen, Zijing Huang, Chuxiao Chen
Cellular Reprogramming.2023; 25(1): 11. CrossRef - Metabolic Remodeling Impacts the Epigenetic Landscape of Dental Mesenchymal Stem Cells
Haiyun Luo, Yachuan Zhou, Wenjing Liu, Jun Wang
Stem Cells International.2022; 2022: 1. CrossRef - SIRT4 regulates rat dental papilla cell differentiation by promoting mitochondrial functions
Haoling Chen, Jun Kang, Fuping Zhang, Tong Yan, Wenguo Fan, Hongwen He, Fang Huang
The International Journal of Biochemistry & Cell Biology.2021; 134: 105962. CrossRef - Sirtuins as Interesting Players in the Course of HIV Infection and Comorbidities
Karolina Jurkowska, Beata Szymańska, Brygida Knysz, Amadeusz Kuźniarski, Agnieszka Piwowar
Cells.2021; 10(10): 2739. CrossRef - Robust expression of SIRT6 inhibits pulpitis via activation of the TRPV1 channel
Jia Hu, Weiran Chen, Zailing Qiu, Hongbing Lv
Cell Biochemistry and Function.2020; 38(5): 676. CrossRef - Downregulation of microRNA‐143‐5p is required for the promotion of odontoblasts differentiation of human dental pulp stem cells through the activation of the mitogen‐activated protein kinases 14‐dependent p38 mitogen‐activated protein kinases signaling pa
Bao‐Liang Wang, Zhi Wang, Xi Nan, Qing‐Cai Zhang, Wei Liu
Journal of Cellular Physiology.2019; 234(4): 4840. CrossRef - A potential role for the silent information regulator 2 homologue 1 (SIRT1) in periapical periodontitis
H. Kudo, O. Takeichi, K. Hatori, K. Makino, K. Himi, B. Ogiso
International Endodontic Journal.2018; 51(7): 747. CrossRef - Overexpressed Sirt1 in MSCs Promotes Dentin Formation in Bmi1-Deficient Mice
H. Wang, C. Lv, Y. Gu, Q. Li, L. Xie, H. Zhang, D. Miao, W. Sun
Journal of Dental Research.2018; 97(12): 1365. CrossRef - Expression of silent information regulator 2 homolog 1 (SIRT1) in periapical granulomas
Hiroshi Kudo, Osamu Takeichi, Kosuke Makino, Keisuke Hatori, Bunnai Ogiso
Journal of Oral Science.2018; 60(3): 411. CrossRef - TET1 knockdown inhibits the odontogenic differentiation potential of human dental pulp cells
Li-Jia Rao, Bai-Cheng Yi, Qi-Meng Li, Qiong Xu
International Journal of Oral Science.2016; 8(2): 110. CrossRef
- Biodegradable Zn‐5Dy Alloy with Enhanced Osteo/Angio‐Genic Activity and Osteointegration Effect via Regulation of SIRT4‐Dependent Mitochondrial Function
- 1,561 View
- 6 Download
- 11 Crossref

- Analysis of gene expression during odontogenic differentiation of cultured human dental pulp cells
- Min-Seock Seo, Kyung-Gyun Hwang, Hyongbum Kim, Seung-Ho Baek
- Restor Dent Endod 2012;37(3):142-148. Published online August 29, 2012
- DOI: https://doi.org/10.5395/rde.2012.37.3.142
-
 Abstract
Abstract
 PDF
PDF PubReader
PubReader ePub
ePub Objectives We analyzed gene-expression profiles after 14 day odontogenic induction of human dental pulp cells (DPCs) using a DNA microarray and sought candidate genes possibly associated with mineralization.
Materials and Methods Induced human dental pulp cells were obtained by culturing DPCs in odontogenic induction medium (OM) for 14 day. Cells exposed to normal culture medium were used as controls. Total RNA was extracted from cells and analyzed by microarray analysis and the key results were confirmed selectively by reverse-transcriptase polymerase chain reaction (RT-PCR). We also performed a gene set enrichment analysis (GSEA) of the microarray data.
Results Six hundred and five genes among the 47,320 probes on the BeadChip differed by a factor of more than two-fold in the induced cells. Of these, 217 genes were upregulated, and 388 were down-regulated. GSEA revealed that in the induced cells, genes implicated in Apoptosis and Signaling by wingless MMTV integration (Wnt) were significantly upregulated.
Conclusions Genes implicated in Apoptosis and Signaling by Wnt are highly connected to the differentiation of dental pulp cells into odontoblast.
-
Citations
Citations to this article as recorded by- iPSC-derived cranial neural crest-like cells can replicate dental pulp tissue with the aid of angiogenic hydrogel
Yoshifumi Kobayashi, Julie Nouet, Erdenechimeg Baljinnyam, Zain Siddiqui, Daniel H. Fine, Diego Fraidenraich, Vivek A. Kumar, Emi Shimizu
Bioactive Materials.2022; 14: 290. CrossRef - The role of sclerostin and dickkopf-1 in oral tissues – A review from the perspective of the dental disciplines
Mohammad Samiei, Klara Janjić, Barbara Cvikl, Andreas Moritz, Hermann Agis
F1000Research.2019; 8: 128. CrossRef - The Influence of Pro-Inflammatory Factors on Sclerostin and Dickkopf-1 Production in Human Dental Pulp Cells Under Hypoxic Conditions
Klara Janjić, Mohammad Samiei, Andreas Moritz, Hermann Agis
Frontiers in Bioengineering and Biotechnology.2019;[Epub] CrossRef - Do hypoxia and L-mimosine modulate sclerostin and dickkopf-1 production in human dental pulp-derived cells? Insights from monolayer, spheroid and tooth slice cultures
Klara Janjić, Barbara Cvikl, Christoph Kurzmann, Andreas Moritz, Hermann Agis
BMC Oral Health.2018;[Epub] CrossRef - The effects of dexamethasone on the apoptosis and osteogenic differentiation of human periodontal ligament cells
Sung-Mi Kim, Yong-Gun Kim, Jin-Woo Park, Jae-Mok Lee, Jo-Young Suh
Journal of Periodontal & Implant Science.2013; 43(4): 168. CrossRef
- iPSC-derived cranial neural crest-like cells can replicate dental pulp tissue with the aid of angiogenic hydrogel
- 1,567 View
- 3 Download
- 5 Crossref

- Gene expression profiling in human dental pulp cells treated with mineral trioxide aggregate
- Yong-Beom Kim, Won-Jun Shon, WooCheol Lee, Kee-Yeon Kum, Seung-Ho Baek, Kwang-Shik Bae
- J Korean Acad Conserv Dent 2010;35(3):152-163. Published online May 31, 2010
- DOI: https://doi.org/10.5395/JKACD.2010.35.3.152
-
 Abstract
Abstract
 PDF
PDF PubReader
PubReader ePub
ePub This study investigated the changes in gene expression when mineral trioxide aggregate (MTA) was applied
in vitro to human dental pulp cells (HDPCs). MTA in a teflon tube (diameter 10 mm, height 2 mm) was applied to HDPCs. Empty tube-applied HDPCs were used as negative control. For microarray analysis, total RNA was extracted at 6, 24, and 72 hrs after MTA application. The results were confirmed selectively by performing reverse transcriptase polymerase chain reaction for genes that showed changes of more than two-fold or less than half. Of the 24,546 genes, 109 genes were up-regulated greater than two-fold (e.g., FOSB, THBS1, BHLHB2, EDN1, IL11, FN1, COL10A1, and TUFT1) and 69 genes were down-regulated below 50% (e.g., SMAD6 and DCN). These results suggest that MTA, rather than being a bio-inert material, may have potential to affect the proliferation and differentiation of pulp cells in various ways.-
Citations
Citations to this article as recorded by- Analysis of gene expression during odontogenic differentiation of cultured human dental pulp cells
Min-Seock Seo, Kyung-Gyun Hwang, Hyongbum Kim, Seung-Ho Baek
Restorative Dentistry & Endodontics.2012; 37(3): 142. CrossRef - Comparison of gene expression profiles of human dental pulp cells treated with mineral trioxide aggregate and calcium hydroxide
Yong-Beom Kim, Won-Jun Shon, Woocheol Lee, Kee-Yeon Kum, Seung-Ho Baek, Kwang-Shik Bae
Journal of Korean Academy of Conservative Dentistry.2011; 36(5): 397. CrossRef
- Analysis of gene expression during odontogenic differentiation of cultured human dental pulp cells
- 1,037 View
- 5 Download
- 2 Crossref

- The comparison of gene expression from human dental pulp cells and periodontal ligament cells
- Hyoun So, Sang-Hyuk Park, Gi-Woon Choi
- J Korean Acad Conserv Dent 2009;34(5):430-441. Published online September 30, 2009
- DOI: https://doi.org/10.5395/JKACD.2009.34.5.430
-
 Abstract
Abstract
 PDF
PDF PubReader
PubReader ePub
ePub The purpose of this study was to characterize functional distinction between human dental pulp cells(PC) and periodontal ligament cells(PDLC) using cDNA microarray assay and to confirm the results of the microarray assay using RT-PCR. 3 genes out of 51 genes which were found to be more expressed(>2 fold) in PC were selected, and 3 genes out of 19 genes which were found to be more expressed(>2 fold) in PDLC were selected for RT-PCR as well.
According to this study, the results were as follows:
1. From the microarray assay, 51 genes were more expressed (2 fold) from PC than PDLC.
2. RT-PCR confirmed that ITGA4 and TGF β2 were more expressed in PC than in PDLC.
3. From the microarray assay, 19 genes were more expressed (2 fold) from PDLC than PC.
4. RT-PCR confirmed that LUM, WISP1, and MMP1 were more expressed in PDLC than in PC.
From the present study, different expression of the genes between the PC and PDLC were characterized to show the genes which play an important role in dentinogenesis were more expressed from PC than PDLC, while the genes which were related with collagen synthesis were more expressed from PDLC than PC.
-
Citations
Citations to this article as recorded by- Gene expression profiling in human dental pulp cells treated with mineral trioxide aggregate
Yong-Beom Kim, Won-Jun Shon, WooCheol Lee, Kee-Yeon Kum, Seung-Ho Baek, Kwang-Shik Bae
Journal of Korean Academy of Conservative Dentistry.2010; 35(3): 152. CrossRef
- Gene expression profiling in human dental pulp cells treated with mineral trioxide aggregate
- 970 View
- 1 Download
- 1 Crossref


 KACD
KACD

 First
First Prev
Prev