Abstract
-
Objectives
We analyzed gene-expression profiles after 14 day odontogenic induction of human dental pulp cells (DPCs) using a DNA microarray and sought candidate genes possibly associated with mineralization.
-
Materials and Methods
Induced human dental pulp cells were obtained by culturing DPCs in odontogenic induction medium (OM) for 14 day. Cells exposed to normal culture medium were used as controls. Total RNA was extracted from cells and analyzed by microarray analysis and the key results were confirmed selectively by reverse-transcriptase polymerase chain reaction (RT-PCR). We also performed a gene set enrichment analysis (GSEA) of the microarray data.
-
Results
Six hundred and five genes among the 47,320 probes on the BeadChip differed by a factor of more than two-fold in the induced cells. Of these, 217 genes were upregulated, and 388 were down-regulated. GSEA revealed that in the induced cells, genes implicated in Apoptosis and Signaling by wingless MMTV integration (Wnt) were significantly upregulated.
-
Conclusions
Genes implicated in Apoptosis and Signaling by Wnt are highly connected to the differentiation of dental pulp cells into odontoblast.
-
Keywords: Differentiation; Gene set enrichment analysis; Human dental pulp cell; Microarray
Introduction
During the tooth morphogenesis, dentin is formed by reciprocal interaction between epithelial cells and dental papilla cells.
1,
2 Some dental papilla cells are differentiated to odontoblast, undifferentiated mesenchymal cell and fibroblast.
3,
4 Undifferentiated mesenchymal cell which is present in pulp space is called dental pulp stem cell because it can be differentiated into odontoblast.
5 Cultured dental pulp cell has self-renewal capacity, multi-lineage differentiation capacity to various organ, and clonogenic efficiency.
6 Dental pulp stem cell can be a useful resource for stem cell therapy including self-stem cell transplantation and tissue regeneration.
7 Gronthos et al. compared renewal capacity of dental pulp stem cell and human bone marrow stromal cells which is known as progenitor of osteoblast.
8 They reported that pulp-like tissue formed dentin-like structure which was surrounded by odontoblast-like cell after injection of dental pulp stem cell into rat.
Many studies using cDNA microarray assay have been done: comparative studies between healthy pulp and caries infected pulp, comparative studies of tooth and bone marrow cells, comparative studies of fibroblast from healthy gingiva and inflammatory gingiva and gene expression comparison between osteoblast and fibroblast.
9-
12 The development of microarray assay for large-scale analysis of mRNA gene expression makes it possible to search key molecules systemincally.
13,
14 After introduction of these genome-wide research techniques, there have been various attempts to describe and compare the gene expression patterns of more specialized adult stem cells for cell characterization.
15,
16 Recently, microarray assay also has been used for investigating the effect of pulp capping materials on human dental pulp cells.
17,
18 Shin et al. reported that at the stage of mineralization of human periodontal ligament (PDL) cells, apoptosis-inducing agents were up-regulated, and anti-apoptosis activators were down-regulated.
19 Even though there are many studies about the differentiation and mineralization of human dental pulp cells, the analyses of the gene expressed in each stage during differentiation are not well understood and remains an active area of investigation.
Therefore, in the present study, we had used a culture system that facilitated the formations of mineralized nodules in human dental pulp cells. We analyzed gene-expression profiles on day 14 of culture using a DNA microarray and sought candidate genes possibly associated with mineralization in an established line of human dental pulp cells characterized by the ability to be differentiated into odontoblastic lineages.
Materials and Methods
Human dental pulp cell isolation
Normal human impacted third molars were collected from 4 healthy adults at the Hanyang University Hospital with informed consent under a protocol approved by the Institutional Review Board of Hanyang University. The tooth surfaces were cleaned and cut around the cementoenamel junction using sterilized diamond stones. Carefully extracted pulp tissue was minced into small pieces and digested in a solution of 3 mg/mL collagenase type 1 (Sigma, St Louis, MO, USA) and 5 mg/mL dispase (Stem cell Technologies, Vancouver, Canada) for 10 minutes at 37℃. Cell suspensions were cultured in 6 cm dishes in high-glucose Dulbecco's modified Eagle's medium (DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and antibiotics (100 U/mL penicillin and 100 mg/mL streptomycin) at 37℃, in 5% CO2.
Odontogenic differentiation in vitro
To induce odontogenic differentiation, dental pulp cells (DPCs) at passage four were seeded in 6 well plates at a density of 1 × 105/well. Odontogenic differentiation was performed for 14 days by using odontoinduction medium (OM) containing α-MEM, 15% FBS, 10 mmol/L β-glycerophosphate (Sigma), 0.2 mmol/L ascorbate-2-phosphated (Sigma), and 100 nmol/L dexamethasone (Sigma). Controls ('non-induced') were cultured in complete medium alone. The media were changed every 3 days.
Evaluation of odontogenic differentiation
To assess in vitro mineralization, cells were washed twice with phosphate buffered saline (PBS), fixed with 4% paraformaldehyde (Sigma) for 1 hour, washed with deionized H2O and stained with 1% Alizarin Red-S (Sigma) for 20 minutes. They were then rinsed three times with deionized H2O and mineralized nodules were visualized and photographed.
The antigen profiles were analyzed by flow cytometric detection of the expression of the stem cell surface markers STRO-1 and CD146. Cells were harvested from induced and non-induced cells with 0.25% trypsin-EDTA. For each analysis, 106 cells/tube were first Fc-blocked with 1 µg of human IgG for 10 minutes and then incubated with mouse antihuman STRO-1 antibody (R&D Systems, Minneapolis, MN, USA) for 1 hour at 4℃. They were washed and centrifuged in 2 mL fluorescence activated cell sorting (FACS) buffer at 10,000 rpm and the supernatants removed, followed by staining with fluorescein isothiocyanate (FITC) conjugated anti-mouse IgM antibody and PE-conjugated mouse antihuman CD146 antibody (R&D systems) for 1 hour at 4℃ in the dark. Residual antibodies were removed by centrifugation and the cells were analyzed with a FACS Canto Flow cytometer (BD Biosciences, San Jose, CA, USA) and FACS DIVA software v6.1.3 (BD Biosciences). All experiments were performed at least three times.
Microarray
Total RNA was extracted using Trizol (Invitrogen) and purified on RNeasy columns (Qiagen, Valencia, CA, USA). After DNase digestion and clean-up procedures, RNA concentrations were measured, and the RNA was aliquoted, and stored at -80℃. RNA purity and integrity were evaluated by denaturing gel electrophoresis, optical density 260/280 ratio, and analysis on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
Total RNA was amplified and purified using an AmbionIllumina RNA amplification kit (Ambion, Austin, TX, USA) to yield biotinylated cRNA. Briefly, 550 ng of total RNA was reverse-transcribed to cDNA using a T7 oligo (dT) primer. Second-strand cDNA was synthesized, in vitro transcribed, and labeled with biotin-Nucleotide Tri-Phosphate (NTP). After purification, the cRNA was quantified with an ND-1000 Spectrophotometer (NanoDrop, Wilmington, DE, USA). 750 ng of labeled cRNA was hybridized to each human HT-12 expression v.4 bead array (Illumina, Inc., San Diego, CA, USA) for 16 - 18 hours at 58℃. Array signals were detected using Amershamfluorolink streptavidin-Cy3 (GE Healthcare Bio-Sciences, Little Chalfont, UK) and scanned with an Illumina bead array reader confocal scanner (Illumina, Inc).
The quality of hybridization and overall chip performance was monitored by visual inspection of both internal quality control checks and the raw scanned data. Raw data were extracted using Gene Expression Module v1.5.4.
Array data were filtered by detecting p values of less than 0.05 for at least 50% of the samples. The selected gene signal values were transformed to logarithms and normalized by the quantile method (n = 6). Samples were compared using fold-change data. All data analyses and visualization procedures for the differentially expressed genes were conducted using ArrayAssist (Stratagene, La Jolla, CA, USA) and R statistical language v. 2.4.1.
Gene set enrichment analysis (GSEA)
Reverse transcription polymerase chain reaction (RT-PCR) analysis
To validate the DNA microarray data, some genes were selected, and their expression was measured by RT-PCR. The RNA was converted to cDNA with Maxime RT-PreMix (Intron, Suwon, Korea) and PCR was also done with Maxime PCR Premix. The target DNA was then amplified by 30 cycles of PCR for all genes using the specific primer pairs in
Table 1, and the PCR products were resolved on 1.5% agarose gels and stained with ethidium bromide. Band intensities corresponding to the target gene and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were measured by image analysis with Chemi-Doc (Bio-Rad, Hercules, CA, USA). The relative amounts of the target mRNA transcripts in the samples were calculated from the ratio of the band intensities of the target genes to that of GAPDH. The analyses were performed in triplicates.
Results
Evaluation of Odontogenic Differentiation
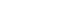
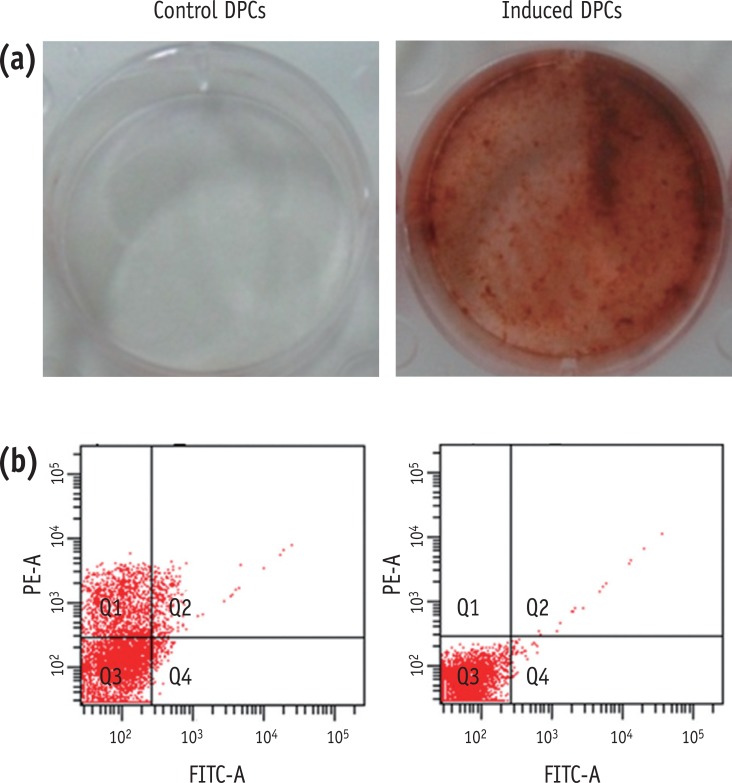
To investigate the hard tissue producing potential of human DPCs undergoing odontogenic differentiation in OM, we assessed Alizarin-Red staining, and levels of transcripts of differentiation markers. Mineralization nodule formation was seen in the DPCs incubated in OM for 14 days (
Figure 1a). To determine the effects of OM on the stem cell properties of DPCs, we tested cells for the stem cell markers, STRO-1 and CD 146. Flow cytometric analysis revealed that the OM-treated cells expressed very much reduced levels of CD146 (0.03%) and STRO-1 (0.7%) (
Figure 1b). This may suggest that the OM-treated DPCs contain a low proportion of stem cells.
Six hundred and five genes among the 47,320 probes on the BeadChip differed by a factor of more than two-fold in the odontogenic induced cells compared with the control cells. Of these, 217 genes were up-regulated, and 388 were down-regulated. Among these genes, those that showed differences of more than five-fold are listed in
Tables 2 and
3. The fold value shows the fold difference between the induced cells and control cells (induced group value/control group value). Positive fold changes refer to upregulation and negative fold changes to down-regulation of gene expression.
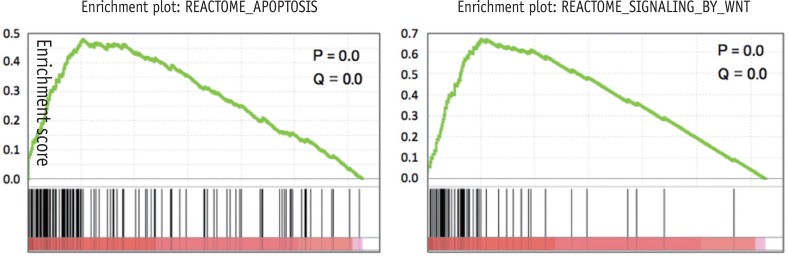
GSEA revealed that odontogenic induction significantly upregulated genes implicated in Apoptosis and Signaling by wingless MMTV integration (Wnt) (
p < 0.01) (
Figure 2).
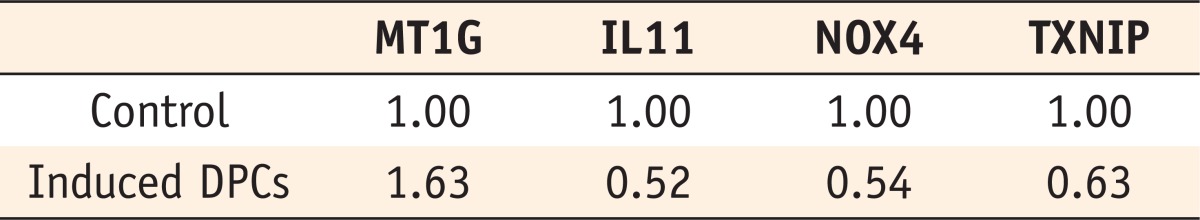
The microarray data were validated using RT-PCR for the following genes: metallothionein 1G (MT1G), interleukin 11 (IL11), thioredoxin interacting protein (TXNIP), and NADPH oxidase 4 (NOX4). These genes are the ones among those showing changes of more than 5-fold that are known to be related to mineralization. The results of the RT-PCR analysis are shown in
Table 4.
Discussion
Couble et al. reported that human dental pulp cells were able to differentiate
in vitro into mature odontoblasts, as assessed by both morphological and functional criteria.
21 They also suggested that such an
in vitro model system is convenient for the study of the mechanisms of odontoblast differentiation and the cellular events associated with extracellular matrix-mediated mineralization. In this study, after 14 days induction, we had confirmed that dishes were fully stained by Alizarlin-Red staining and stemness of dental pulp cells was almost lost by FACS results. This means that stem cells which were present in the cultured dental pulp cells had been already differentiated into odontoblast-like cells by odontogenic induction procedure.
Microarray results showed that many genes were changed in the course of odontogenic differentiation. Among those genes, genes that showed more than 5 times and specially related to osteoblast differentiation was MT1G, NOX-4, IL11, and TXNIP. In considering the role of these 4 genes, changes of these genes in this study are matched with osteoblast differentiation. MT-1 and MT-2 are acute-phase proteins co-induced in response to various agents such as heavy metals, glucocorticoids, oxidative stress, and cytokines, and their individual functions are not clear. Miyahara et al. observed that undifferentiated osteoblasts synthesized MT on exposure to dexamethasone with enhanced calcification in the presence of β-glycerophosphate.
22 Furthermore, Lin et al. suggested that ZnCl
2 is able to promote dental pulp stem cells (DPSCs) differentiation by upregulating MT.
23 In our study, MT1G was significantly increased after odontogenic induction. BMP-2 is necessary and sufficient to induce the differentiation of stem cells into odontoblasts and BMP-2 protein production is modulated by ROS generated by NOX-4.
24,
25 IL-11 expression plays an important role in the stimulation of osteoblast differentiation and increased IL-11 enhances osteoblastogenesis by stimulating Wnt/β-catenin signaling.
26 In this study, NOX-4 and IL-11 were significantly down-regulated after odontogenic induction. It is well-documented that ROS induces TXNIP expression and consequently induces osteoclast differentiation. In this study, TXNIP was significantly down-regulated after odontogenic induction.
We used GSEA to explore novel pathways regulated differentially by odontogenic induction. As many biological processes occur as a network of changes in the expression levels of many genes, the analysis of a few genes may not be sufficient to predict biological phenomena. However, it is difficult to analyze biological processes that are distributed across entire networks of genes, and affected by subtle changes at the level of individual genes. GSEA revealed that odontogenic induction upregulated gene sets involved in Apoptosis and Signaling by Wnt. Wnt proteins have been recently introduced as core regulators of bone synthesis. The canonical pathway that works through intra-cellular transducer β-catenin, control differentiation of osteoblast progenitor cells into mature osteoblasts. β-catenin is expressed in mesenchymal precursor cells and its inactivation favors their differentiation into chondrocytes instead of osteoblast.
27 Human receptor mutations that inactivates Wnt signal results in a generalized increase in bone mass.
28 Wnt signaling regulates osteoblast differentiation by adjusting several transcription factors including Runx2. Wnt signal promotes Runx2 expression and activity. LEF/TCF transcription factors, the end point of Wnt signal in the nucleus promotes Runx2 and Osterix expression and interacts with Runx2 to control its function during osteoblast differentiation.
29 Shin et al. reported that at the stage of mineralization of human PDL cells, apoptosis-inducing agents were up-regulated, and anti-apoptosis activators were down-regulated.
19 In this study, we confirmed that apoptosis is important in the mineralization event not only in human PDL cells but also in human dental pulp cells. Therefore, we concluded that genes implicated in Apoptosis and Signaling by Wnt are highly connected to the differentiation of dental pulp cells into odontoblast.
Conclusions
In this study, we conclude that genes implicated in Apoptosis and Signaling by Wnt are highly connected to the differentiation of dental pulp cells into odontoblast. However, by fragmentary comparing two different cells, we cannot conclude exactly which gene is more responsible for a certain role. More scientific verifications and studies are needed for that purpose.
-
This study was supported by Biostart Program from graduate school of biomedical science and engineering, Hanyang University.
-
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1. Ruch JV. In: Linde A, editor. Tooth morphogenesis and differentiation. Dentin and Dentinogenesis. 1984. Boca Raton: CRC press; p. 47-79.
- 2. Kikuchi H, Sawada T, Yanagisawa T. Effects of a functional agar surface on in vitro dentinogenesis induced in proteolytically isolated agar-coated dental papillae in rat mandibular incisors. Arch Oral Biol 1996;41:871-883.ArticlePubMed
- 3. Ruch JV. Odontoblast differentiation and the formation of the odontoblast layer. J Dent Res 1985;64:489-498.ArticlePubMedPDF
- 4. Osman M, Ruch JV. Behavior of odontoblasts and basal lamina of trypsin or EDTA-isolated mouse dental papillae in short-term culture. J Dent Res 1981;60:1015-1027.ArticlePubMedPDF
- 5. Ruch JV. Odontoblast commitment and differentiation. Biochem Cell Biol 1998;76:923-938.ArticlePubMed
- 6. Gronthos S, Brahim J, Li W, Fisher LW, Cherman N, Boyde A, DenBesten P, Robey PG, Shi S. Stem cell properties of human dental pulp stem cells. J Dent Res 2002;81:531-535.ArticlePubMedPDF
- 7. Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, Shi S. SHED: Stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA 2003;100:5807-5812.ArticlePubMedPMC
- 8. Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA 2000;97:13625-13630.ArticlePubMedPMC
- 9. Pääkkönen V, Ohlmeier S, Bermann U, Larmas M, Salo T, Tjäderhane L. Analysis of gene and protein expression in healthy and carious tooth pulp with cDNA microarray and two-dimensional gel electrophoresis. Eur J Oral Sci 2005;113:369-379.ArticlePubMed
- 10. Shi S, Robey PG, Gronthos S. Comparison of human dental pulp and bone marrow stromal stem cells by cDNA microarray analysis. Bone 2001;29:532-539.ArticlePubMed
- 11. Wang PL, Ohura K, Fujii T, Oido-Mori M, Kowashi Y, Kikuchi M, Suetsugu Y, Tanaka J. DNA microarray analysis of human gingival fibroblasts from healthy and inflammatory gingival tissues. Biochem Biophys Res Commun 2003;305:970-973.ArticlePubMed
- 12. Han X, Bolcato AL, Amar S. Identification of genes differentially expressed in cultured human osteoblasts versus human fibroblasts by DNA microarray analysis. Connect Tissue Res 2002;43:63-75.ArticlePubMed
- 13. Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet 1999;21:10-14.ArticlePubMedPDF
- 14. Brown PO, Bostein D. Exploring the new world of the genome with DNA microarrays. Nat Genet 1999;21:33-37.ArticlePubMedPDF
- 15. Inomata N, Tomita H, Ikezawa Z, Saito H. Differential gene expression profile between cord blood progenitorderived and adult progenitor-derived human mast cells. Immunol Lett 2005;98:265-271.ArticlePubMed
- 16. Schilling T, Küffner R, Klein-Hitpass L, Zimmer R, Jakob F, Schütze N. Microarray analyses of transdifferentiated mesenchymal stem cells. J Cell Biochem 2008;103:413-433.ArticlePubMed
- 17. Kim YB, Shon WJ, Lee W, Kum KY, Baek SH, Bae KS. Comparison of gene expression profiles of human dental pulp cells treated with mineral trioxide aggregate and calcium hydroxide. J Korean Acad Conserv Dent 2011;36:397-408.Article
- 18. Kim YB, Shon WJ, Lee W, Kum KY, Baek SH, Bae KS. Gene expression profiling in human dental pulp cells treated with mineral trioxide aggregate. J Korean Acad Conserv Dent 2010;35:152-163.Article
- 19. Shin JH, Park JW, Yeo SI, Noh WC, Kim MK, Kim JC, Suh JY. Identification of matrix mineralization-related genes in human periodontal ligament cells using cDNA microarray. J Korean Acad Periodontol 2007;37:447-463.ArticlePDF
- 20. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545-15550.ArticlePubMedPMC
- 21. Couble ML, Farges JC, Bleicher F, Perrat-Mabillon B, Boudeulle M, Magloire H. Odontoblast differentiation of human dental pulp cells in explant cultures. Calcif Tissue Int 2000;66:129-138.ArticlePubMedPDF
- 22. Miyahara T, Nemoto M, Tukamoto S, Yamada H, Kozuka H, Kuze S, Sudo H, Yamamoto S. Induction of metallothionein and stimulation of calcification by dexamethasone in cultured clonal osteogenic cells, MC3T3-E1. Toxicol Lett 1991;57:257-267.ArticlePubMed
- 23. Lin CY, Lin HH, Tsai MH, Lin SP, Chen MH. Zinc Chloride for odontogenesis of dental pulp stem cells via metallothionein up-regulation. J Endod 2011;37:211-216.ArticlePubMed
- 24. Casagrande L, Demarco FF, Zhang Z, Araujo FB, Shi S, Nör JE. Dentin-derived BMP-2 and odontoblast differentiation. J Dent Res 2010;89:603-608.ArticlePubMedPDF
- 25. Mandal CC, Ganapathy S, Gorin Y, Mahadev K, Block K, Abboud HE, Harris SE, Ghosh-Choudhury G, Ghosh-Choudhury N. Reactive oxygen species derived from Nox4 mediate BMP2 gene transcription and osteoblast differentiation. Biochem J 2011;433:393-402.ArticlePubMedPDF
- 26. Kido S, Kuriwaka-Kido R, Imamura T, Ito Y, Inoue D, Matsumoto T. Mechanical stress induces Interleukin-11 expression to stimulate osteoblast differentiation. Bone 2009;45:1125-1132.ArticlePubMed
- 27. Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell 2005;8:739-750.PubMed
- 28. Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med 2002;346:1513-1521.ArticlePubMed
- 29. Bennett CN, Longo KA, Wright WS, Suva LJ, Lane TF, Hankenson KD, MacDougald OA. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci USA 2005;102:3324-3329.ArticlePubMedPMC
Figure 1(a) Left: Dishes of control (non-induced) DPCs, Right: induced (OM-treated) DPCs after Alizarin-Red staining; (b) The expression of surface markers as analyzed by flow cytometry of control and induced DPCs. Left: control DPCs. Right: induced DPCs. Q3 means cells exhibits negative to STRO-1 and CD146 antibody. DPC, dental pulp cell; OM, odontogenic induction medium.
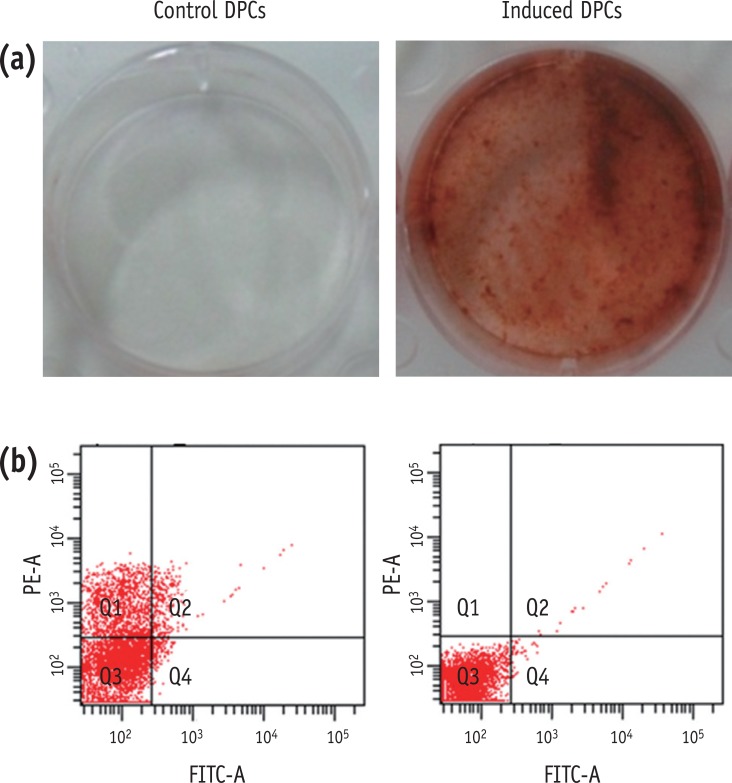
Figure 2Gene set enrichment analysis (GSEA) identifying novel pathways activated in odontogenic inducted cells relative to the control cells.
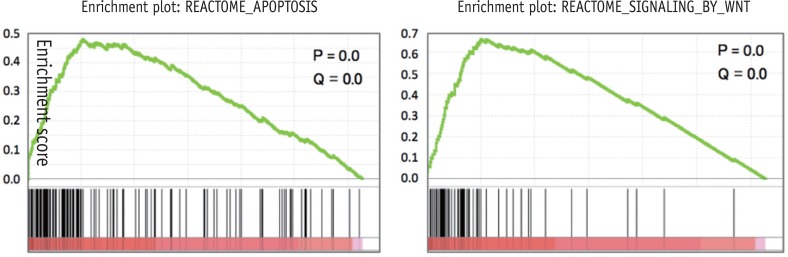
Table 1
Table 2Genes up-regulated on day 14 in odontogenic medium (over a five-fold change)
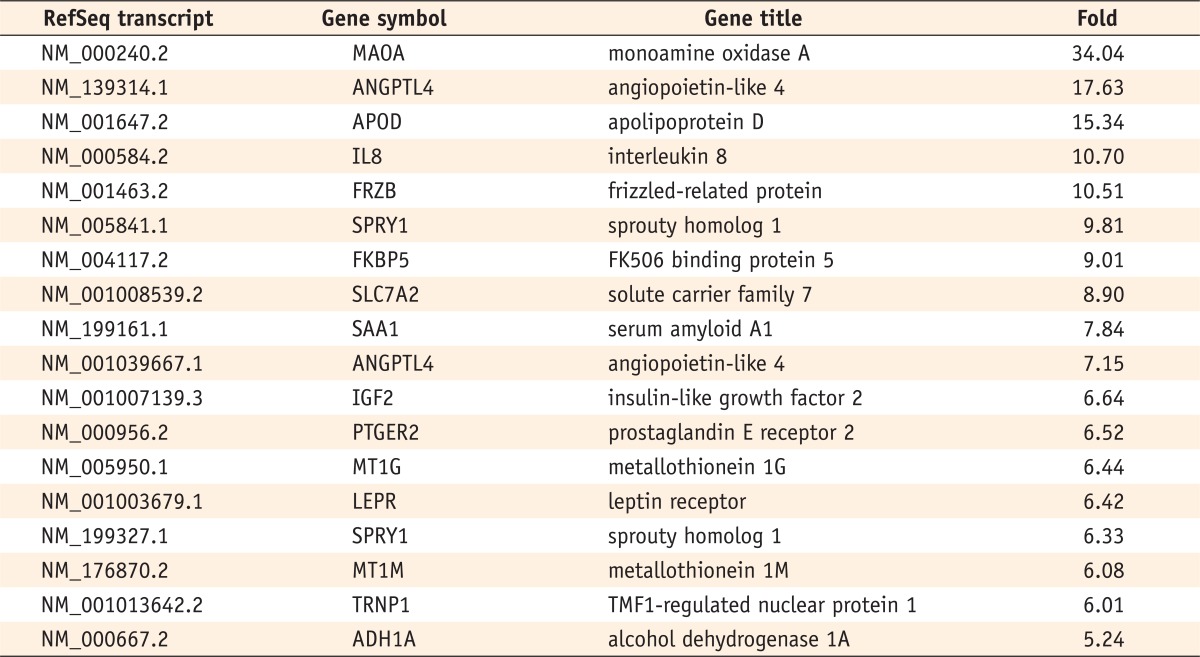
Table 3Genes down-regulated on day 14 in odontogenic medium (over a five-fold change)
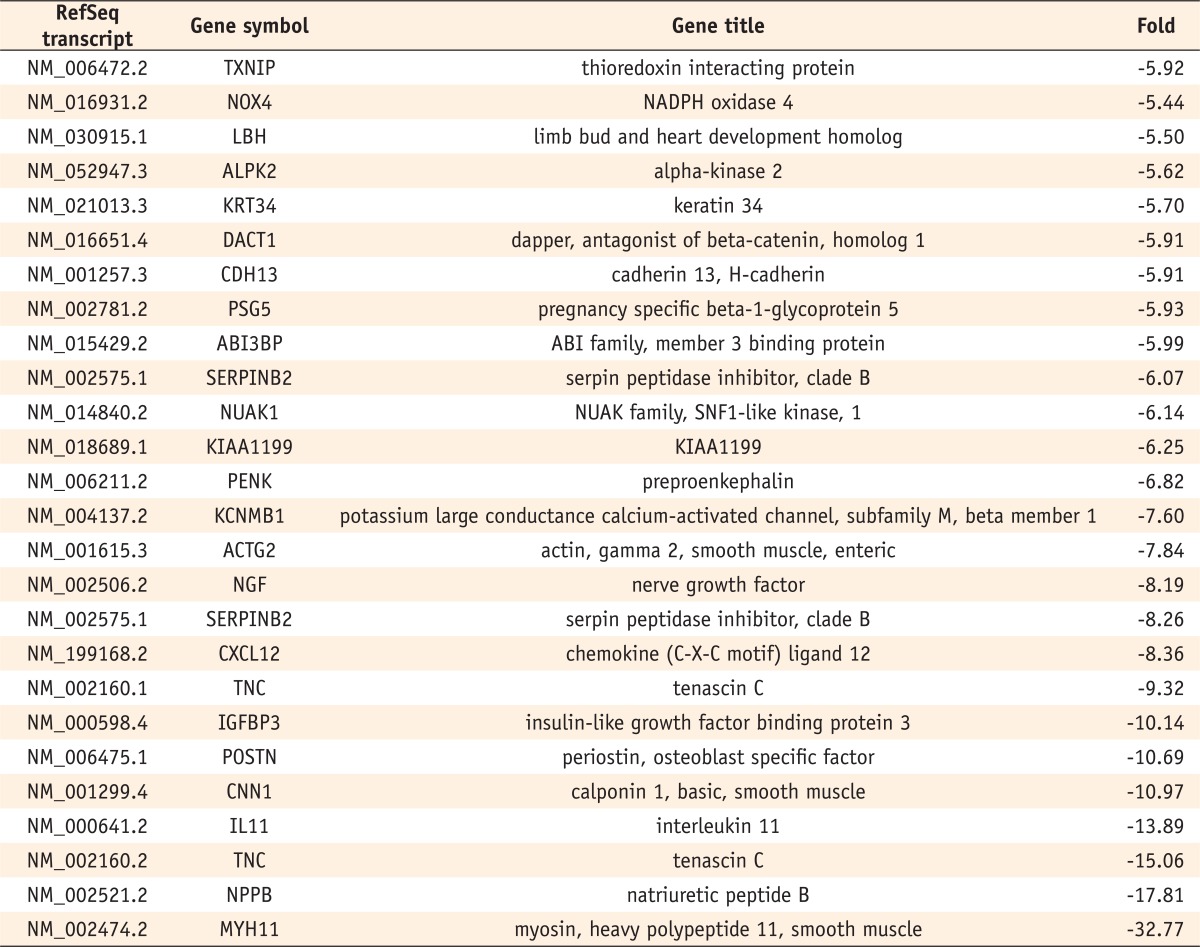
Table 4Four genes selected for semi-quantitative RT-PCR analysis. Relative levels of gene expression are normalized against GAPDH messenger RNA and each control is set at 1.0










 KACD
KACD


 ePub Link
ePub Link Cite
Cite